Vertebrate Immune Responses Involve Communication

The Study of Microbe–Host Ii-Fashion Communication
Section of Beast and Food Sciences, University of Delaware, Newark, DE 19716, USA
*
Author to whom correspondence should be addressed.
Bookish Editor: Mohammad Katouli
Received: 15 Dec 2021 / Revised: 2 February 2022 / Accustomed: seven February 2022 / Published: x February 2022
Abstruse
Dorsum-and-forth intercommunication in host–pathogen interactions has long been recognized to play an important role in commensalism and microbial pathogenesis. For centuries, we accept studied these microbes in our surroundings, yet many questions nearly the evolutionary cross-talk between host and microbe remain unanswered. With the recent surge in enquiry interest in the commensal microbiome, basic immunological questions have returned to the fore, such equally, how are vast numbers of microbes capable of coexisting within animals and humans while also maintaining a healthy functional immune system? How is the evasion and subversion of the immune organisation achieved by some microbes but not others? The intricate and of import-to-remember two-way interaction and coevolution of host and microbe is the communication network we must tap into equally researchers to answer these questions.
1. The Host Immune System
The vertebrate allowed system is classified into 2 main branches, the innate and adaptive immune systems [1,2]. The innate immune system is the start line of defense against strange invaders; therefore, innate allowed cells are equipped with special "generalist" receptors that recognize molecular patterns plant on microbes, known as pattern recognition receptors (PRRs) [3]. PRRs on immune cells bind to microbes, or components of microbes, and trigger allowed responses. This recognition is not microbe specific and relies on the recognition motifs independent in a variety of microbes rather than a discriminating antigen. The adaptive allowed arrangement is evolutionarily more advanced and is highly discriminating between microbes. In the case of bacteria, when elimination of bacteria is not carried out by the innate PRR induced activities, both bacteria lysis and phagocytosis can be induced past opsonization facilitated past the adaptive allowed system equally well other adaptive responses [4,five]. Complement mediated lysis of invasive bacteria outside the cell requires the binding of specific antibodies to the target leaner to initiate a complex capable of invading and disrupting the bacterial cell. These antibodies are produced past B-cells of the adaptive immune system. Phagocytosis can also be initiated by the binding of antibodies and acute stage proteins such as C-reactive protein and serum amyloid A and P to allow presentation on major histocompatibility complex (MHC)-II of macrophages. The binding of T-prison cell Receptor (TCR) to MHC-II stimulates the production of cytokines [3] thus, inducing intracellular leaner killing mechanisms such as lysozyme activity.
There are a number of studies that describe bacterially induced changes in host allowed signaling [six,7,8,9,x,xi,12]. Every bit mentioned earlier, one of the host's first lines of defence force against bacteria is via activation of PRRs. There are three main PRRs involved in the emptying of bacteria by the innate immune arrangement; these are Toll-similar receptors (TLRs), nucleotide-binding and oligomerization (NOD)- like receptors (NLRs), and C-blazon lectin receptors (CLRs) [3]. Following the activation of these receptors, there is an increase in signal transduction cascades that involve mitogen-activated poly peptide kinase (MAPK), interferon (IFN), nuclear Factor kappa-light-chain enhancer of activated B cells (NFkB)—these are other signaling proteins that induce inflammatory responses and clear the bacteria. These cascades are heavily dependent on protein kinases for activation and regulation. Subsequently, nosotros will discuss how these kinases can get targets for microbes to influence the host immune response.
2. Bacterial Response to the Host
A big proportion of immune inquiry related to host immune–pathogen interaction focuses on the adverse outcomes during its (dys)function or how it can be manipulated when in a diseased land. This is understandable as researchers are attempting to study infections of importance with regard to disease. Notwithstanding, understanding the optimum functioning of the immune organisation in the context of a readily eliminated infectious threat (rather than a serious or chronic affliction) may help usa empathise the bones processes of the allowed system and its optimum response to microbes. For researchers studying host–pathogen interactions that result in affliction states the question arises, what dynamic are nosotros studying, an efficient immune mobilization to address an infectious agent, or a microbial beneficial response designed to evade the immune response? For instance, Salmonella induces inflammatory responses at the epithelial layer of the gut, using the resultant permeability to invade across the gut bulwark [9,13]. While the inflammatory response destroys many leaner, it besides allows invasive Salmonella to take concur inside the host. This invasion across the endothelium is disquisitional for the survival and spread of Salmonella within the host. Although it has been shown that Salmonella can survive in the lumen [fourteen,15,16], it is a hostile environs for long-term survival. Some species harbor Salmonella, tolerate its presence and it tin can become part of the commensal microflora, thus facilitating its continuous shedding [17]. In other species, such as humans, the inflammatory allowed responses induced by Salmonella triggers diarrhea which clears the contents of the lumen including nutrients, debris and bacteria [14] seems to necessitate invasion. The ability of Salmonella to exploit the inflammatory allowed response is beneficial and critical for its survival. Infection of the epithelial or immune cells that line the lumen may effect in disruption of the epithelial bulwark due to the inflammation triggered by the presence of the bacteria [16], which facilitates their spread to the lamina propria. Moreover, to successfully infect organs outside of the gastrointestinal (GI) tract, Salmonella must cross the endothelial bulwark and travel through the bloodstream. This is benign to the bacteria considering colonizing different organs is optimal for growth and survival. This as well poses a threat considering crossing the endothelium into the blood causes septicemia and other fatal health complications especially in immunocompromised hosts. Who is responding to whom in this state of affairs? Mayhap the answer to such questions eludes us because information technology is difficult to study the immune arrangement when information technology is functioning ordinarily and properly, or when it is in a homeostatic remainder with a microbial population.
3. The Case of Leaner Kinases
Host immune responses to bacteria vary partly considering of the many different changes in signal transduction that can occur during exposure to pathogenic and commensal leaner. Bacteria are capable of inducing changes not just in the host gene expression but also in the proteome past straight or indirect modification of proteins [18,19]. Here, nosotros will focus on one instance, the post-translational modification (PTM) via phosphorylation of the host proteome by bacterial kinases. Phosphorylation is the add-on of a gamma phosphate from an adenosine triphosphate (ATP) molecule to a specific serine, threonine or tyrosine amino acid rest of protein [xx,21,22,23,24]. This covalent process is catalyzed by enzymes known as kinases, kinases are found in both eukaryotes and prokaryotes. The word kinase is derived from the Greek word Kinein which ways to move [24]. Kinases are phosphotransferases thus they move phosphate groups from 1 organic molecule to some other, this is not limited to simply proteins. Kinases tin can be classified into canonical kinases or pseudokinases [25]. Canonical refers to kinases that are catalytically active and pseudokinases refers to those with an evolutionary loss of function. That is, pseudokinases are inactive kinases that evolved alongside catalytically active kinases but lack cardinal requirements to serve equally kinases [26]. Research suggests that pseudokinases may play an of import function in regulating other kinases [27]. Pseudokinases are also common to both eukaryotes and prokaryotes. Biologically disquisitional kinases known to be involved in regulatory cellular processes such metabolism, allowed regulation, cell maintenance, etc. are part of a big superfamily of canonical protein kinases. In eukaryotes, canonical protein kinases are divided into 2 main subfamilies, namely: the protein-serine/threonine kinases (STKs) and the protein-tyrosine kinases [22]. These kinases serve as on and off switches for many cellular processes. The protein-serine/threonine kinases are either membrane spring or intracellular signaling proteins that phosphorylate the oxygen of a hydroxyl (OH) group on serine or threonine amino acids. Examples of protein-serine/threonine kinases include MAPK kinases, protein kinase (A, B, C), Casein kinase, calcium/calmodulin kinases, etc. The protein-tyrosine kinases (PTKs) are either transmembrane or cytoplasmic poly peptide kinases that phosphorylate the tyrosine residuum of a protein. PTKs include receptor tyrosine kinases such as epithelial growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), vascular endothelial growth factor receptor (VEGFR) and non-receptor tyrosine kinases such as janus kinase (JAK), SRC, SYK family.
Kinases homologous and orthologous to the STK subfamily are found in one of the four main classes of bacterial kinases [28]. The form of bacterial kinases similar to STKs are called eukaryotic-similar serine/threonine kinases (eSTKs). eSTKs are involved in a plethora of activities disquisitional to the survival of the bacteria in a host, including the modulation of host proteome, bacterial cell wall synthesis, bacterial metabolism, and others [five] (Cozzone 2005). Studies take been shown that Salmonella and Yersinia eSTKs tin can straight dispense hosts actin and cytoskeletal activity via MAP, myosin and Rho/Rac kinase activities which can alter processes such equally phagocytosis, cell growth and vesicle germination in the host [nine,10,29]. Kinases of M. tuberculosis are capable of altering metabolism activities in host cells by inhibiting the regulation of tricarboxylic acid (TCA) cycle enzymes [8,10]. Kinases in this bacteria have also been shown to subtract host inflammatory response and increment bacteria loads [7,30,31]. Some other class of bacteria kinases of annotation is leaner tyrosine (By) kinase. By kinases are kinases that phosphorylate tyrosine residues in bacteria and are non homologous to eukaryote Hanks-type tyrosine kinases, however, further studies are required to prove orthology [32,33]. The ability of manipulating kinase part in health and disease has been known in human medicine for decades, a massive research and evolution enterprise worth many billions of dollars has focused on the inhibition of kinase action in the treatment of cancers [34,35,36]. The fact that bacteria have taken advantage of the host use of kinase-mediated signal transduction to facilitate invasion and persistence is an excellent example of microbe–host coevolution.
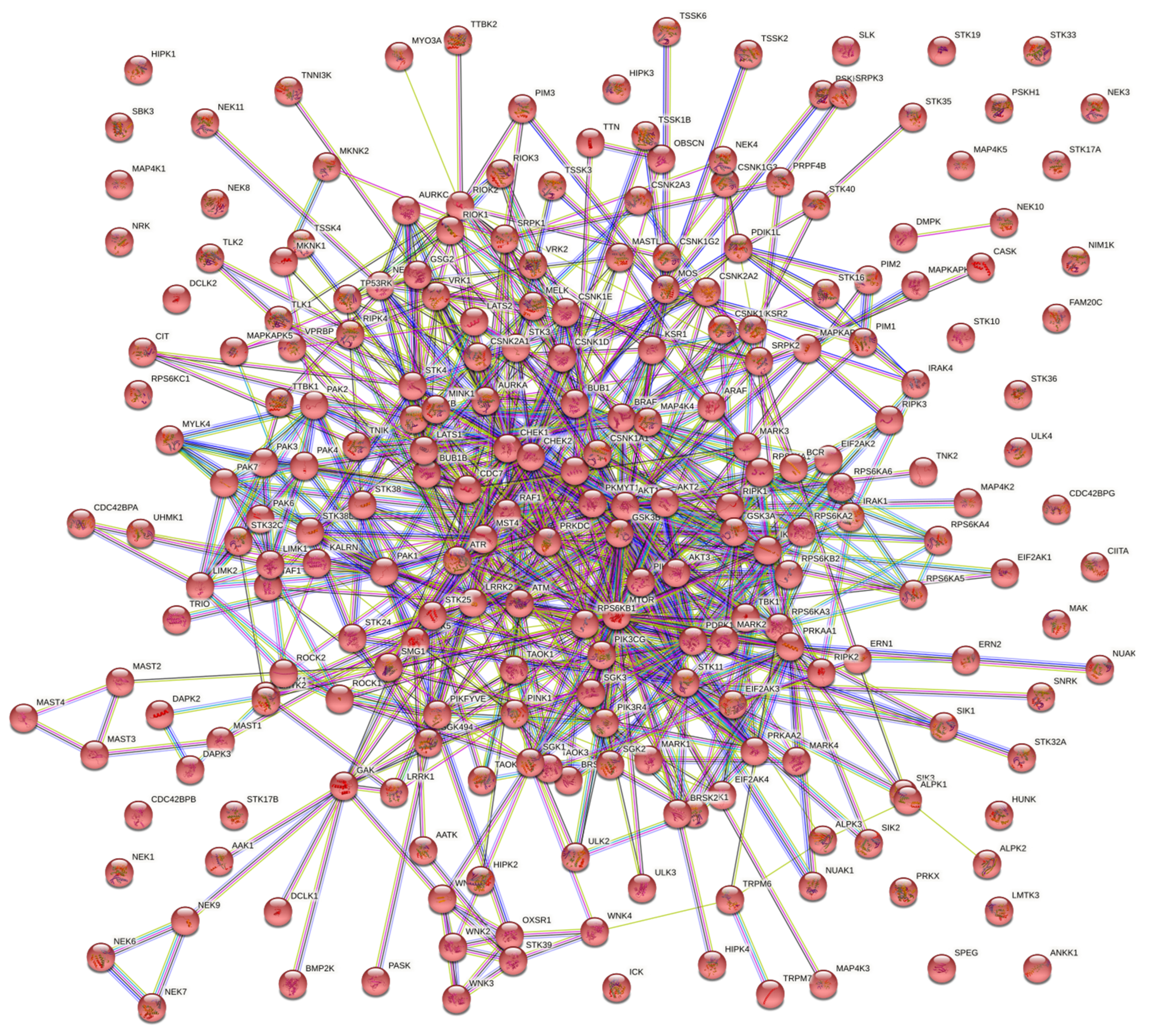
To emphasize the importance of kinases further, a uniprotKB based curation of human protein kinases that play a office in immune response resulted in 234 reviewed entries (Figure 1). Among these 234 kinases, there are over 200 also involved in metabolism, illustrating the indisputable importance of immunometabolism in fully understanding microbe–host interactions. The results of this curation suggests the affluence of targets of opportunity available to bacteria just in the context of kinases capable of influencing the immune organization.
4. Microbe–Host Crosstalk and Commensals
Pathogenic leaner interactions with the host take been studied in more than particular than commensal bacteria. Thus more is understood about their kinase action on hosts than commensal bacteria, despite commensals' obvious benefits and intriguing effects on the immune system. Balance and homeostasis are central to a mutually beneficial coexistence between the microbiota and host. In the host, the microbiota finds a secure surround with favorable conditions such as temperature, oxygen levels and nutrient availability. The microbiota through its activities improves host metabolism, for example, by producing vitamins, and increasing nutrient availability by improving digestibility of circuitous carbohydrates [39,40]. A well-established microbiota competitively excludes pathogens by outcompeting pathogens for nutrients and an ecological niche within the host. The commensal microbiota provides critically important signals to the immune system for its development and maturation The abundance of microbes, mostly bacteria that reside in the intestinal tract, is evident of the complex relationship between the microbiota and the host immune organisation [41,42,43]. The microbiota tin be harmful to the host if bacteria cantankerous the epithelial bulwark or if not-commensal microbes' entry into the intestinal lumen is left unchecked. Changes caused by life events tin can disrupt the microbial niche and result in dysbiosis which may favor pathogens. Too changes in metabolic benefits as discussed above, such disruption of the microbiome may curb the tolerogenic signals the microbiome produces, triggering proinflammatory responses that disrupt allowed homeostasis. Such adverse events have been linked to diseases in many systems of hosts' bodies other than the gut, namely, diabetes, atopic dermatitis, multiple sclerosis, asthma, etc. [44,45,46].
An instance of an allowed response with nuanced cross-talk between host and microbe is that of secretory immunoglobulin A (sIgA). sIgAs in the mucosal lining are of import antibodies for initiating immune responses and for the competitive inhibition of leaner binding to epithelial cells in the mucosa [47]. Study results regarding the specificity of sIgA to eliminate commensal compared to pathogenic leaner differ. Many studies show that sIgA prevents overstimulation of the immune system to answer to bacteria that may be beneficial to the host [48]. Transportation of commensal bacteria across the Peyer'southward patch M cells and dendritic cells is proposed to be essential in the host tolerance of these bacteria [49]. Tolerance is also promoted by a sIgA-commensal complex in the mucosa [48]. The exact mechanism by which the vertebrate allowed system is capable of fostering such a relationship is however to exist determined. Mathias and Corthésy showed that removal of glycans found on sIgA significantly decreased its interaction with Gram-positive bacteria, indicating modification of sIgA can also alter the fate of the bacteria and the specificity of sIgA [50].
It is clear that during homeostasis, the host immune system does not reply to commensal bacteria, or certainly not in the same fashion every bit it does to pathogenic or exogenous bacteria. Perhaps, this is a consequence of a sort of symbiotic coevolution betwixt the microbiota and its hosts resulting in a permanent modify in physiology to facilitate tolerance of the microbiota [eighteen]. Researchers estimate that bacteria take been circumstantial with vertebrates for approximately 0.5 billion years; this dynamic interaction has shaped the microbial community and immune system, and immune the tolerance of the microbiota by the allowed system [51]. The history of innate immune cells' evolution of PRR tin can be traced dorsum to Cnidarians (invertebrates) need for a sort of immune specificity that distinguished symbionts from pathogens [52]. The development of the adaptive immune system corresponds to the evolution of vertebrates [53]. The oldest vertebrates need to obtain antigen recognition receptors on T-cells and B-cells as well equally long lived allowed memory allowed precise identification of and appropriate response to a vast majority of antigens non only as an upgrade to the host defense force but more importantly, to distinguish between friends and foes, similar to their invertebrate predecessors.
five. Conclusions
The data provided in this newspaper argues that our approach to agreement the two-way communication between host and microbe in terms of health and diseases is lacking. This also highlights the difficulty associated with agreement bacteria populations where differences tin exist down to the serovars of specific species [12]. The written report of infectious disease is a study of both the immune response to self and the, sometimes, inappropriate or co-opted immune response. Further exploration of the immunometabolic benefits offered by coexisting with specific microbes is needed. The sequence of events that triggered the tolerance and fostering of microbial niches are yet to be understood, but the benefits of such events are undeniable. Understanding these mechanisms may let us to care for a wide variety of infectious diseases (viral, bacterial, fungal, parasitic) as well every bit chronic inflammatory diseases (for example, Crohn's, lupus, and arthritis).
Author Contributions
Conceptualization, writing—original draft preparation, writing—review and editing, F.P. and R.J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Involvement
The authors declare no conflict of interest.
References
- Boehm, T. Evolution of vertebrate immunity. Curr. Biol. 2012, 22, R722–R732. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The Innate and Adaptive Immune Systems. Constitute for Quality and Efficiency in Wellness Care (IQWiG). Available online: https://www.ncbi.nlm.nih.gov/books/NBK279396/ (accessed on 15 December 2021).
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, G.J. Receptors of the Innate Allowed System. Immunobiology: The Immune Organization in Wellness and Affliction fifth edition. 2001. Available online: https://world wide web.ncbi.nlm.nih.gov/books/NBK27129/ (accessed on 2 December 2021).
- Merle, Due north.Southward.; Noe, R.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement system role II: Role in immunity. Front. Immunol. 2015, 6, 257. [Google Scholar] [CrossRef] [PubMed][Dark-green Version]
- Thau, L.; Asuka, Eastward.; Mahajan, K. Physiology, Opsonization; StatPearls Publishing: Treasure Island, FL, United states of america, 2021. Bachelor online: http://world wide web.ncbi.nlm.nih.gov/books/NBK534215/ (accessed on fifteen December 2021).
- Cozzone, A.J. Role of poly peptide phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J. Mol. Microbiol. Biotechnol. 2005, 9, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Gupta, Thou.; Pathak, M.; Gupta, N.; Koul, A.; Sarangi, S.; Baweja, R.; Singh, Y. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, embr, in vivo. J. Bacteriol. 2006, 188, 2936–2944. [Google Scholar] [CrossRef] [PubMed][Light-green Version]
- O'Hare, H.Thou.; Durán, R.; Cerveñansky, C.; Bellinzoni, Thousand.; Wehenkel, A.One thousand.; Pritsch, O.; Obal, G.; Baumgartner, J.; Vialaret, J.; Johnsson, Yard.; et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 2008, lxx, 1408–1423. [Google Scholar] [CrossRef] [PubMed]
- Odendall, C.; Rolhion, Northward.; Förster, A.; Poh, J.; Lamont, D.J.; Liu, Grand.; Freemont, P.S.; Catling, A.D.; Holden, D.W. The Salmonella Kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Jail cell Host Microbe 2012, 12, 657–668. [Google Scholar] [CrossRef][Green Version]
- Canova, M.J.; Baronian, G.; Brelle, South.; Cohen-Gonsaud, M.; Bischoff, Thou.; Molle, V. A novel manner of regulation of the Staphylococcus aureus Vancomycin-resistance-associated response regulator VraR mediated by Stk1 poly peptide phosphorylation. Biochem. Biophys. Res. Commun. 2014, 447, 165–171. [Google Scholar] [CrossRef]
- Pagano, G.J.; Arsenault, R.J. Advances, challenges and tools in characterizing bacterial serine, threonine and tyrosine kinases and phosphorylation target sites. Practiced Rev. Proteom. 2019, 16, 431–441. [Google Scholar] [CrossRef]
- Perry, F.; Johnson, C.; Aylward, B.; Arsenault, R.J. The differential phosphorylation-dependent signaling and glucose immunometabolic responses induced during infection by salmonella enteritidis and salmonella heidelberg in craven macrophage-like cells. Microorganisms 2020, 8, 1041. [Google Scholar] [CrossRef]
- Poh, J.; Odendall, C.; Spanos, A.; Boyle, C.; Liu, M.; Freemont, P.; Holden, D.Due west. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Jail cell Microbiol. 2008, 10, 20–30. [Google Scholar] [CrossRef][Green Version]
- Santos, R.L.; Raffatellu, M.; Bevins, C.L.; Adams, L.K.; Tükel, Ç.; Tsolis, R.One thousand.; Bäumler, A.J. Life in the inflamed intestine, Salmonella mode. Trends Microbiol. 2009, 17, 498–506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loetscher, Y.; Wieser, A.; Lengefeld, J.; Kaiser, P.; Schubert, South.; Heikenwalder, M.; Hardt, W.D.; Stecher, B. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS ONE 2012, 7, e34812. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luk, C.H.; Valenzuela, C.; Gil, Thousand.; Swistak, L.; Bomme, P.; Chang, Y.Y.; Mallet, A.; Enninga, J. Salmonella enters a dormant state inside homo epithelial cells for persistent infection. PLoS Pathog. 2021, 17, e1009550. [Google Scholar] [CrossRef] [PubMed]
- Kogut, Grand.H.; Arsenault, R.J. A part for the not-canonical Wnt-β-catenin and TGF-β signaling pathways in the induction of tolerance during the establishment of a Salmonella enterica serovar enteritidis persistent cecal infection in chickens. Front. Vet. Sci. 2015, ii, 33. [Google Scholar] [CrossRef][Dark-green Version]
- Tran van Nhieu, G.; Arbibe, L. Genetic reprogramming of host cells by bacterial pathogens. F1000 Biol. Rep. 2009, 1, eighty. [Google Scholar] [CrossRef][Greenish Version]
- Bonne Køhler, J.; Jers, C.; Senissar, M.; Shi, L.; Derouiche, A.; Mijakovic, I. Importance of protein Ser/Thr/Tyr phosphorylation for bacterial pathogenesis. FEBS Lett. 2020, 594, 2339–2369. [Google Scholar] [CrossRef][Greenish Version]
- Krebs, East.Chiliad.; Fischer, East.H. Phosphorylase activeness of skeletal muscle extracts. J. Biol. Chem. 1955, 216, 113–120. [Google Scholar] [CrossRef]
- Krebs, E.Thou. The phosphorylation of proteins: A major mechanism for biological regulation. Biochem. Soc. Trans. 1985, 13, 813–820. [Google Scholar] [CrossRef][Dark-green Version]
- Hanks, S.K.; Hunter, T. Poly peptide kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Cohen, P. The origins of protein phosphorylation. Nat. Prison cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef]
- Lehrer, S.; Bloomer, W.D. Activity of pp6oc-src protein kinase in man breast cancer. Mt. Sinai J. Med. 1989, 56, 83–85. [Google Scholar] [PubMed]
- Shaw, A.Southward.; Kornev, A.P.; Hu, J.; Ahuja, Fifty.1000.; Taylor, S.Due south. Kinases and Pseudokinases: Lessons from RAF. Molecular and Cellular Biology. 2014. Bachelor online: https://journals.asm.org/doi/abs/10.1128/MCB.00057-14 (accessed on 15 Dec 2021).
- Raju, S.; Shaw, A.Southward. What is the point of pseudokinases? eLife 2015, 4, e07771. [Google Scholar] [CrossRef] [PubMed]
- Rajakulendran, T.; Sicheri, F. Allosteric protein kinase regulation by pseudokinases: Insights from STRAD. Sci. Signal 2010, 3, pe8. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, Northward.; Anamika, Yard.; Srinivasan, N. A Framework for nomenclature of prokaryotic protein kinases. PLoS ONE 2010, five, e10608. [Google Scholar] [CrossRef]
- Lee, Westward.L.; Grimes, J.M.; Robinson, R.C. Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization. Nat. Struct. Mol. Biol. 2015, 22, 248–255. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Briken, V.; Porcelli, South.A.; Besra, 1000.Due south.; Kremer, L. Mycobacterial lipoarabinomannan and related lipoglycans: From biogenesis to modulation of the immune response. Mol. Microbiol. 2004, 53, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Papavinasasundaram, 1000.G.; Chan, B.; Chung, J.-H.; Colston, M.J.; Davis, E.O.; Av-Gay, Y. Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J. Bacteriol. 2005, 187, 5751–5760. [Google Scholar] [CrossRef][Light-green Version]
- Jadeau, F.; Bechet, Eastward.; Cozzone, A.J.; Deléage, M.; Grangeasse, C.; Combet, C. Identification of the idiosyncratic bacterial protein tyrosine kinase (By-kinase) family signature. Bioinformatics 2008, 24, 2427–2430. [Google Scholar] [CrossRef][Light-green Version]
- Grangeasse, C.; Nessler, S.; Mijakovic, I. Bacterial tyrosine kinases: Evolution, biological role and structural insights. Philos. Trans. R. Soc. B. Biol. Sci. 2012, 367, 2640–2655. [Google Scholar] [CrossRef][Green Version]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.G.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and futurity directions. Mol. Cancer. 2018, 17, 48. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.One thousand.; Brunak, South.; Campbell, A.; Gan, 1000.N.; Gaulton, A.; Gomez, Due south.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery xx years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, xx, 551–569. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, G.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-poly peptide clan networks with increased coverage, supporting functional discovery in genome-broad experimental datasets. Nucleic. Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rowland, I.; Gibson, M.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed][Greenish Version]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Part of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef][Dark-green Version]
- Wu, H.-J.; Wu, E. The role of gut microbiota in allowed homeostasis and autoimmunity. Gut Microbes 2012, three, 4–xiv. [Google Scholar] [CrossRef][Greenish Version]
- Belkaid, Y.; Hand, T. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef][Green Version]
- Tomkovich, S.; Jobin, C. Microbiota and host allowed responses: A beloved–detest relationship. Immunology 2016, 147, 1–10. [Google Scholar] [CrossRef][Green Version]
- Hassenbach, Thou.; Albus, One thousand. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema. Lancet 1998, 351, 1225–1232. [Google Scholar]
- Bach, J.-F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002, 347, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.-F. The "hygiene hypothesis" for autoimmune and allergic diseases: An update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Buckland, J. Behaviour of secretory IgA explained. Nat. Rev. Immunol. 2003, 3, 519. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA's complex roles in amnesty and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef]
- Favre, L.; Spertini, F.; Corthésy, B. Secretory IgA possesses intrinsic modulatory properties stimulating mucosal and systemic immune responses. J. Immunol. 2005, 175, 2793–2800. [Google Scholar] [CrossRef][Greenish Version]
- Mathias, A.; Corthésy, B. Recognition of gram-positive intestinal leaner by hybridoma- and colostrum-derived secretory immunoglobulin a is mediated past carbohydrates. J. Biol. Chem. 2011, 286, 17239–17247. [Google Scholar] [CrossRef][Green Version]
- Craig, L.One thousand.; Charles, O.Due east.; Robin, D.H.; Casey, T.W. Reciprocal Interactions of the Abdominal Microbiota and Allowed Organization. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef][Green Version]
- Emery, M.A.; Dimos, B.A.; Mydlarz, L.D. Cnidarian pattern recognition receptor repertoires reverberate both phylogeny and life history traits. Front end. Immunol. 2021, 12, 2430. [Google Scholar] [CrossRef]
- McFall-Ngai, M. Treat the community. Nature 2007, 445, 153. [Google Scholar] [CrossRef]
Figure 1. Network of protein kinases involved in immune responses in humans from STRING-database. Homo poly peptide kinases with functions in immune response were curated using UniprotKB [37]. The resulting 234 review entries were entered into String-database to generate the interaction network [38].
Figure 1. Network of protein kinases involved in immune responses in humans from STRING-database. Homo protein kinases with functions in immune response were curated using UniprotKB [37]. The resulting 234 review entries were entered into String-database to generate the interaction network [38].
| Publisher's Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open admission article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/iv.0/).
Vertebrate Immune Responses Involve Communication,
Source: https://www.mdpi.com/2076-2607/10/2/408/htm
Posted by: jenkinsexchilliked.blogspot.com


0 Response to "Vertebrate Immune Responses Involve Communication"
Post a Comment